Introduction
In this article, we will compare the different ways of loading data into Redshift tables from data present in AWS S3 and HDFS.
- Amazon S3: Amazon S3 (Simple Storage Service)is a cloud IaaS (Infrastructure as a Service) solution from Amazon Web Services for object storage via a convenient web-based interface. According to Amazon, the benefits of S3 include industry-leading scalability, data availability, security, and performance.
- Amazon EMR HDFS: HDFS (Hadoop Distributed File System)is a distributed, scalable, and portable file system for Hadoop. HDFS seeks to provide a distributed, fault-tolerant file system that can run on commodity hardware.

Scenario
- The user uploads a large data file in a compressed format to the S3 bucket using an S3 pre-signed URL.
- An S3 event is pushed to the SQS queue which acts as a trigger for a lambda function.
- The lambda function starts an EMR step to process the input data file.
- EMR processes the raw data and loads the transformed data to redshift.
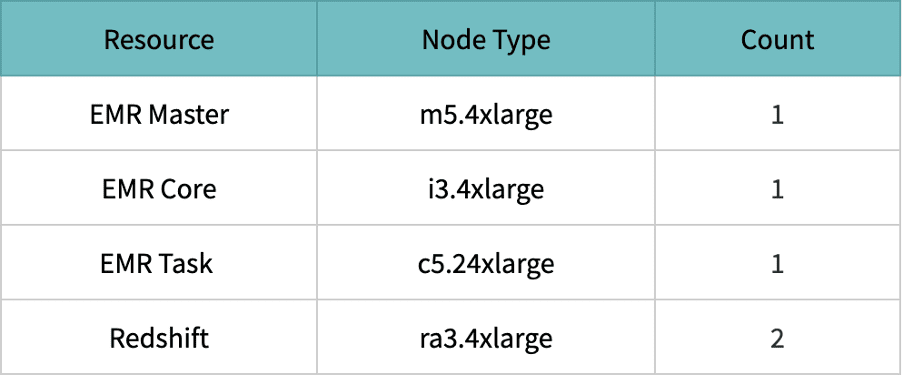
The above scenario is tested with the following resource configurations,
Resource configuration
Input file

Scenario 1: Load data to Redshift using S3 as intermediary store via COPY command
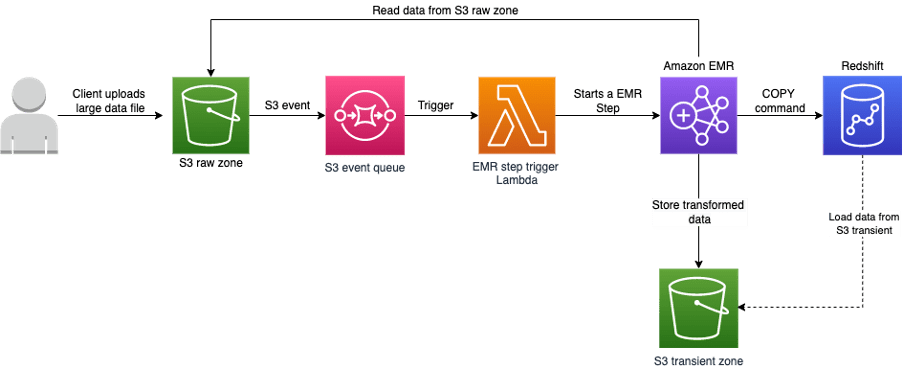
For Loading data from S3 to Redshift, we should satisfy the below prerequisites,
- IAM role associated with the redshift cluster.
- Redshift Security inbound allows EMR connections.
EMR script to load S3 transient data to redshift:
from pyspark.sql import SparkSession
import pyspark.sql.functions as F
import uuid
from sqlalchemy import create_engine
from sqlalchemy.orm import Session
import sqlalchemy
from datetime import datetime
import argparse
# Parsing the input arguments
parser = argparse.ArgumentParser()
parser.add_argument("--app-name", help="Spark bulk application name",)
parser.add_argument("--file-path", help="Input file path",)
parser.add_argument("--out-bucket", help="Output bucket name",)
args = parser.parse_args()
app_name = args.app_name
file_path = args.file_path
bucket = args.out_bucket
# Application Constants
prefix = 'transient/{}/'.format(str(uuid.uuid4()))
temp_dir_path = 's3://{}/{}'.format(bucket, prefix)
author = 'vishal'
null_type = '@NULL@'
output_table_name = 'sample'
output_file_format = "CSV GZIP"
# Config values fetched from secret manager
username = 'username'
password = 'Password'
url = 'Cluster host url'
port = 'Cluster port'
database = 'Cluster database name'
iam_role = 'Cluster associated IAM role'
# Building the spark session
execution_start_time = datetime.now()
spark = SparkSession.builder.appName(app_name).getOrCreate()
spark_context = spark.sparkContext
# Reading input file from S3 path
df_csv = spark.read.format("csv") \
.option("header", True) \
.option("multiLine", True) \
.option("ignoreLeadingWhiteSpace", True) \
.option("ignoreTrailingWhiteSpace", True) \
.option("escape", "\\") \
.option("quote", "\"") \
.load(file_path)
# Performing minimal transformation
modified_df = df_csv.withColumn('load_by', F.lit(author))
# Writing modified dataframe to s3 bucket
modified_df.write \
.format("csv") \
.option("escape", "\"") \
.option("nullValue", null_type) \
.option("compression", "gzip") \
.option("header", True) \
.mode("append").save(temp_dir_path)
# Establishing Redshift connection using sqlalchemy
redshift_url = "postgresql://{}:{}@{}:{}/{}".format(
username, password, url, port, database)
connection_engine = create_engine(
redshift_url, pool_size=0, max_overflow=-1,
pool_pre_ping=True).execution_options(autocommit=True)
connection = Session(bind=connection_engine,
expire_on_commit=False,
autocommit=True).connection()
# Building the COPY command
columns = ','.join(modified_df.columns)
query = "COPY {} ({}) FROM '{}' iam_role '{}' FORMAT AS {} NULL AS '{}'
TIMEFORMAT 'auto' IGNOREHEADER 1;"
final_query = query.format(output_table_name, columns,
temp_dir_path, iam_role, output_file_format,
null_type)
# Executing the COPY command
connection.execute(sqlalchemy.text(final_query))
total_time = str((datetime.now() -
execution_start_time).total_seconds())
print('Total time taken {} seconds'.format(total_time))
Scenario 1: Load data to Redshift using S3 as intermediary store via COPY command
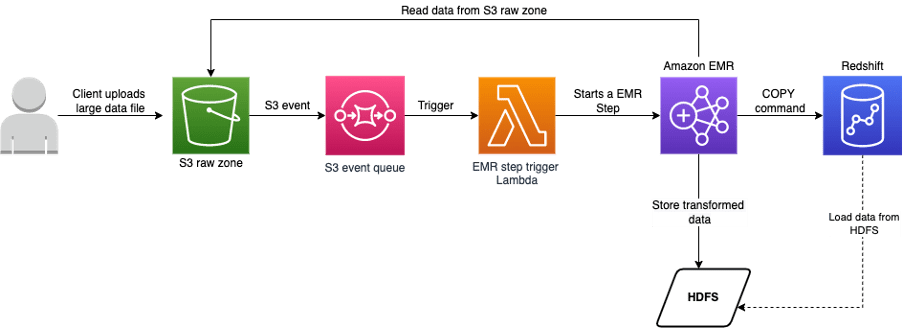
For Loading data from HDFS to Redshift, we should satisfy the below prerequisites,
- Redshift Public Key should be under authorized keys of the EMR cluster.
- IAM role associated with the redshift cluster.
- Redshift Security inbound allows EMR connections.
The EMR bootstrap script would look as follows,
#!/bin/bash
aws redshift describe-clusters --cluster-identifier redshift-cluster |
jq -r '.Clusters[0].ClusterPublicKey' >>
/home/hadoop/.ssh/authorized_keys
set -e
cur_dir=`pwd`
pwd
echo -e "Installing Python dependencies:\n"
sudo echo -e
"boto3==1.17.35\nwheel==0.36.2\nSQLAlchemy==1.4.13\npsycopg2-
binary==2.8.6" >> requirements.txt
sudo cat requirements.txt
sudo easy_install-3.7 pip
sudo /usr/local/bin/pip3 install -r requirements.txt --
target=/usr/local/lib/python3.7/site-packages
The EMR code for loading data from HDFS to redshift would look like follows,
from pyspark.sql import SparkSession
import pyspark.sql.functions as F
import uuid
from sqlalchemy import create_engine
from sqlalchemy.orm import Session
import sqlalchemy
from datetime import datetime
import argparse
# Parsing the input arguments
parser = argparse.ArgumentParser()
parser.add_argument("--app-name", help="Spark bulk application name",)
parser.add_argument("--file-path", help="Input file path",)
args = parser.parse_args()
app_name = args.app_name
file_path = args.file_path
# Application Constants
prefix = 'transient/{}/'.format(str(uuid.uuid4()))
author = 'vishal'
null_type = '@NULL@'
output_table_name = 'sample'
output_file_format = "CSV GZIP"
# Config values fetched from secret manager
username = 'username'
password = 'Password'
url = 'Cluster host url'
port = 'Cluster port'
database = 'Cluster database name'
iam_role = 'Cluster associated IAM role'
# These variables are modified for the HDFS flow.
cluster_id = 'EMR Cluster ID'
temp_dir_path = "hdfs:///{}".format(prefix)
final_path = 'emr://{}/'.format(cluster_id) + prefix + 'part*'
# Building the spark session
execution_start_time = datetime.now()
spark = SparkSession.builder.appName(app_name).getOrCreate()
spark_context = spark.sparkContext
# Reading input file from S3 path
df_csv = spark.read.format("csv") \
.option("header", True) \
.option("multiLine", True) \
.option("ignoreLeadingWhiteSpace", True) \
.option("ignoreTrailingWhiteSpace", True) \
.option("escape", "\\") \
.option("quote", "\"") \
.load(file_path)
# Performing minimal transformation
modified_df = df_csv.withColumn('load_by', F.lit(author))
# Writing modified dataframe to HDFS
modified_df.write \
.format("csv") \
.option("escape", "\"") \
.option("nullValue", null_type) \
.option("compression", "gzip") \
.option("header", True) \
.mode("append").save(temp_dir_path)
# Establishing Redshift connection using sqlalchemy
redshift_url = "postgresql://{}:{}@{}:{}/{}".format(
username, password, url, port, database)
connection_engine = create_engine(
redshift_url, pool_size=0, max_overflow=-1, pool_pre_ping=True).execution_options(autocommit=True)
connection = Session(bind=connection_engine,
expire_on_commit=False, autocommit=True).connection()
# Building the COPY command
columns = ','.join(modified_df.columns)
query = "COPY {} ({}) FROM '{}' iam_role '{}' FORMAT AS {} NULL AS '{}' TIMEFORMAT 'auto' IGNOREHEADER 1;"
final_query = query.format(output_table_name, columns,
final_path, iam_role, output_file_format, null_type)
# Executing the COPY command
connection.execute(sqlalchemy.text(final_query))
total_time = str((datetime.now() - execution_start_time).total_seconds())
print('Total time taken {} seconds'.format(total_time))
In this article, we will compare the different ways of loading data into Redshift tables from data present in AWS S3 and HDFS.
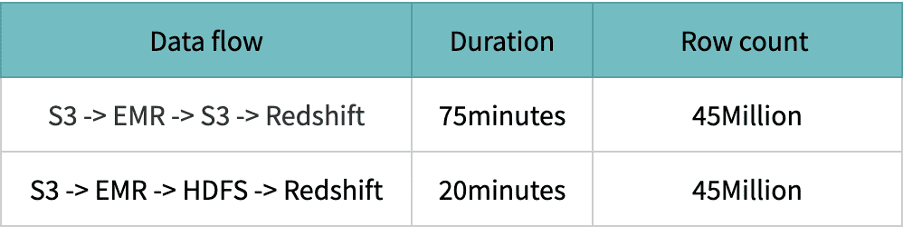
HDFS performance is excellent compared to S3 because the data is stored and processed on the same machines; access and processing speed are lightning-fast.
Reason for low performance on S3
Spark leverages Hadoop’s FileOutputCommitter implementations to write data. Writing data again involves multiple steps and on a high level – staging output files and then committing them i.e. writing final files. Here, the rename step is involved from staging to the final step. In AWS S3 (object store), the file rename under the hood is a Copy followed by a Delete operation. The source file is first copied to the destination and then, it is deleted. The renames process depends on the size of the file. If there is a directory rename, then it depends on the number of files inside the dir along with the size of each file. So in a nutshell, renaming is a very expensive operation in S3 as compared to a normal file system.
Caveat on using HDFS as transient zone
HDFS relies on local storage that scales horizontally. If you want to increase your storage space, you’ll either have to add larger hard drives to existing nodes or add more machines to the cluster. This is feasible but more costly and complicated than S3.
Overcoming the caveat
Once the file is successfully loaded to the redshift table, delete the file from the HDFS transient zone and have a CRON job to frequently monitor the HDFS location. Remove the files older than X amount of period.
Code snippet to schedule a CRON for every 6-hours via EMR bootstrap script.
EMR bootstrap script
#!/bin/bash
sudo aws s3 cp s3://source-bucket/hdfs_cleaner.sh /home/hadoop/poc/
sudo chmod +x /home/hadoop/poc/hdfs_cleaner.sh
sudo crontab<<EOF
0 */6 * * * sh /home/hadoop/poc/hdfs_cleaner.sh
EOF
HDFS cleaner code to delete the files which are older than a day.
EMR HDFS cleaner
#!/bin/bash
hdfs dfs -ls /transient/* |
tr -s " " |
cut -d' ' -f6-8 |
grep "^[0-9]" |
awk '{
cmd = "date +%s";
cmd | getline NOW;
cmd = "date -d'\''"$1" "$2"'\'' +%s";
cmd | getline WHEN;
DIFF = NOW-WHEN;
MIN = 1440;
LAST = 60*MIN;
if(DIFF > LAST){
print "Deleting: " $3;
print "hdfs dfs -rm -r -skipTrash " $3
system("hdfs dfs -rm -r "$3);
}
}';
References
- https://docs.aws.amazon.com/redshift/latest/dg/loading-data-from-emr.html
- https://www.integrate.io/blog/storing-apache-hadoop-data-cloud-hdfs-vs-s3/
- https://routdeepak.medium.com/writing-to-aws-s3-from-spark-91e85d09724b
- https://stackoverflow.com/questions/44235019/delete-files-older-than-10days-on-hdfs